3x10 for 10/23: Evolution, Pooled genetic screens, Immunity and Single cell atlases
3-sentence summaries of the 10 coolest life science papers out in October 2023
Hi, friends!
Welcome to October’s 3x10, our monthly roller-coaster journey through October’s coolest life science papers 🚀🧬. In this edition, you’ll read 3-sentence summaries of papers on evolution, single cell methodologies, genetic screens, and more. At the end of the newsletter, you’ll get an interesting video as a Halloween bonus 👻.
Enjoy 3x10!
Assembly theory (Sharma et al., Nature). The most (in)famous paper I read this month proposes a new framework (assembly theory, that is) to explain basically everything… or, more specifically, “to unify descriptions of evolutionary selection across physics and biology”. This paper is not an easy read for anybody (in particular evolutionary biologists), but, to its merit, it sparked scientific discussions by being different than what is expected for a scientific paper describing evolution. The abstract is particularly philosophical (and arguably tricky at times), and states that assembly theory “discloses a new aspect of physics emerging at the chemical scale, whereby history and causal contingency influence what exists”… food for thought, I guess.
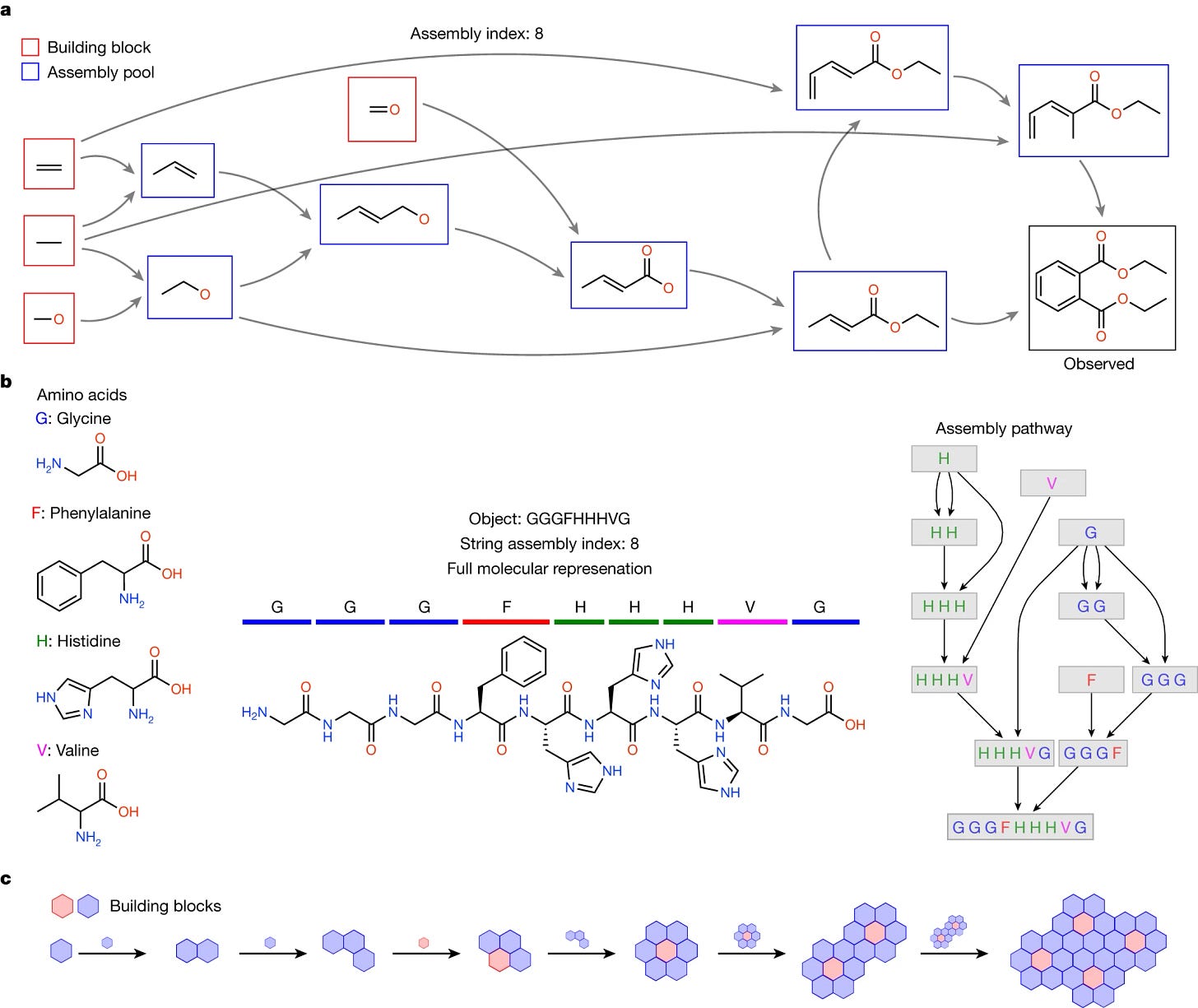
1. Assembly theory explains and quantifies selection and evolution. We don’t really understand what this all means, but it does motivate us to think differently about the theory and evolution of life. Here’s the associated explainer in Nature with an even more artsy title: How purposeless physics underlies purposeful life. Efficient and scalable compressed Perturb-seq (Yao et al., Nature Biotechnology). Perturb-seq is a technique that combines pooled genetic screening via CRISPR with single-cell RNA sequencing to assess the separate impacts of different genetic perturbations on a cell's gene expression profile in a single experiment for a cellular population (aka many CRISPR perturbations + scRNAseq readout, for a population of cells). Using an experimental and computational framework inspired by compressed sensing applied to genetic screens, this study manages to substantially reduce the number of measurements needed to run Perturb-seq, leading to an impressive reduction in experimental cost (and with only small changes to the original protocol). Compressed Perturb-seq is a wonderful example of innovative theory applied to wet-lab experiments with direct economic value, which will hopefully allow increased adoption of Perturb-seq screening.

2. Scalable genetic screening for regulatory circuits using compressed Perturb-seq, an innovative experimental & computational approach applying compressed sensing to genetic screens and achieving substantial cost reductions for Perturb-seq experiments. Population-level integration of single-cell datasets (De Donno et al., Nature Methods): More and more single cell data is being added to single cell atlases, yet it is not clear how to optimally combine existing datasets to preserve biological signal and remove technical artifacts, or how to interact with collections of cells in the context of new data. scPoli is a computational framework for single-cell population level integration of scRNAseq & scATACseq datasets; an open-world learner that incorporates generative models to learn sample and cell representations for data integration, label transfer and reference mapping. The tool (available as an integral part of scArches) is able to rigorously complete a bunch of useful tasks when dealing with atlases: build references from semi-labeled datasets, compare donors, identify novel cell types, extend a reference with new cell types, and even integrate across species.
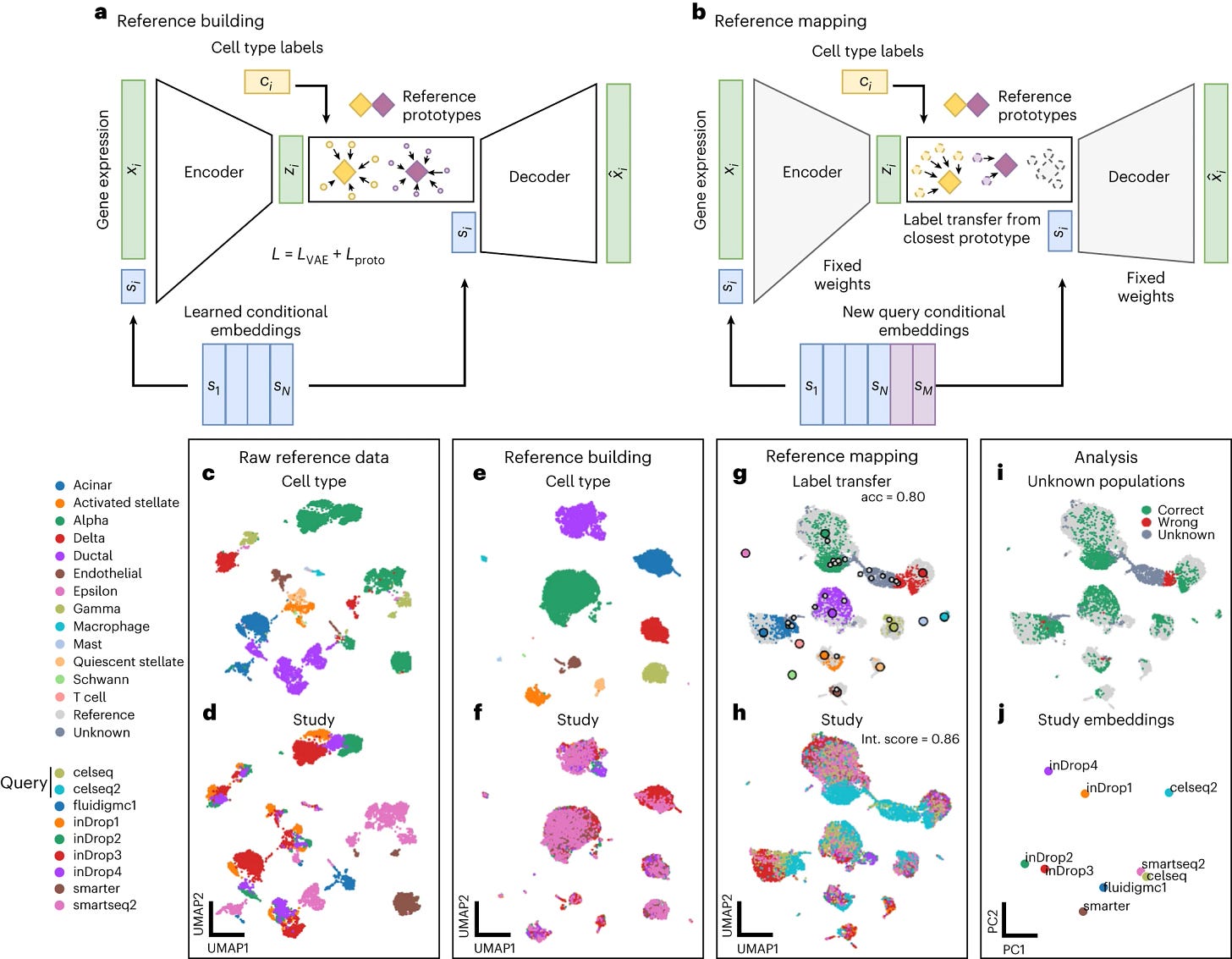
3. Population-level integration of single-cell datasets enables multi-scale analysis across samples, an open-world learner that incorporates generative models to learn sample and cell representations for data integration, label transfer and reference mapping. EVEscape (Thadani et al., Nature). A computational framework that combines fitness predictions from a deep learning model of historical sequences with biophysical and structural information, with the aim of predicting future viral evolution. EVEscape quantifies the viral escape potential of mutations and, by doing so, it manages to accurately predict nearly all known escape variants of SARS-CoV-2 from pre-pandemic sequence data alone. Moving forward, EVEscape can predict probable future SARS-CoV-2 mutations to forecast emerging strains as a tool to inform continuous and rational mRNA vaccine design, and to estimate the pandemic risk for previously unknown viruses.

4. Learning from prepandemic data to forecast viral escape, a very nice and also useful tool to predict future viral emerging strains and aid in rational mRNA vaccine design. Here’s the insightful explainer by Nature. Brain Cell Census (Science). In an impressive scientific effort, 21 papers published simultaneously shine light on the cellular composition and functionality of the most complex, magical and fascinating neural networks ever designed: our own brains. It comes as no surprise that these papers report an immense complexity of the prenatal, postnatal and adult human brain, consisting of thousands of cell types and states, with specific functionality. The papers are a valuable resource for the community and an important step forward in understanding living organisms.
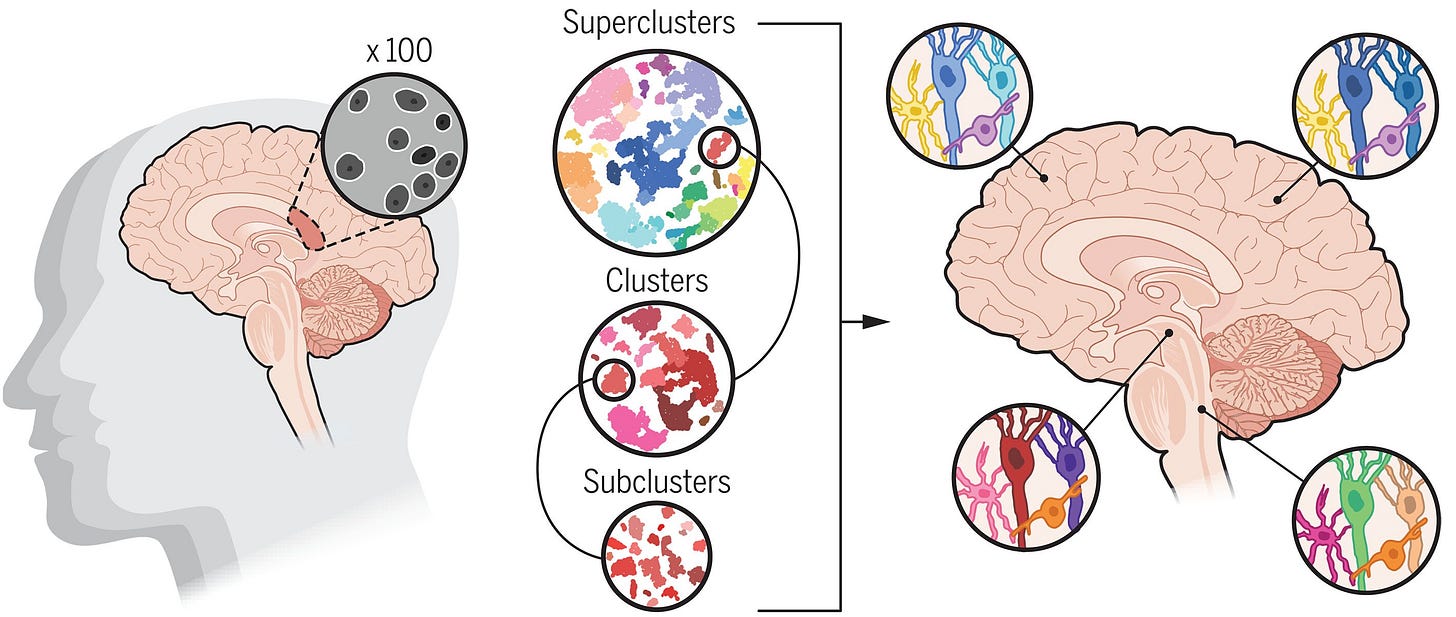
5. Transcriptomic diversity of cell types across the adult human brain (Siletti et al.), one of the main studies in this collection: thorough characterization into functional cell types and states for 2,000,000 neurons from 100 anatomical locations of postmortem brain tissue belonging to 3 donors. Mosaic copy number alterations in normal tissues (Gao et al., Nature Genetics). This paper adds to the literature unravelling the evolution of somatic variations in normal tissues, and its relation to aging and diseases such as cancer. By inferring mosaic copy number alterations (mCAs) from the GTEX (Genotype-Tissue Expression) database of bulk gene expression profiles on normal tissues, the paper concludes that clonal expansions of mCAs are more widespread in normal tissues than previously thought: mCAs were found in more than 20 tissues, a quarter of the subjects harbored an mCA somewhere in the body, and the prevalence of clonal mCAs increased with age, from almost inexistent in people under 30 to 30% in people above 50. The prevalence and patterns of mCAs across tissues suggest tissue-specific mutagenic exposure and selection pressures, mirroring previous findings on the evolution of somatic point mutations in normal tissues.
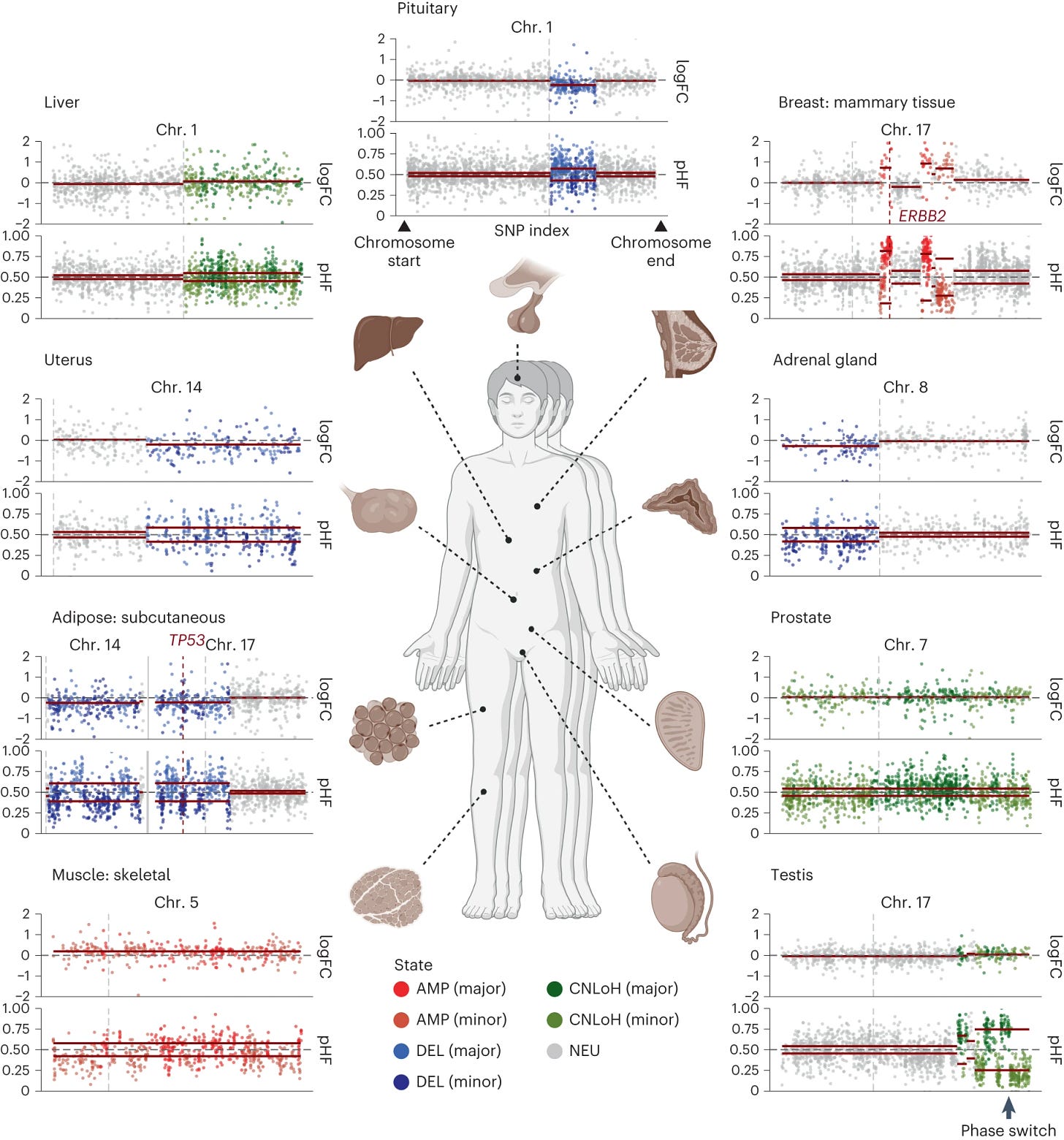
6. A pan-tissue survey of mosaic chromosomal alterations in 948 individuals: a computational approach to infer mosaic copy number alterations in normal tissue from expression profiles. P.S.: paper #5 covered in September’s 3x10 edition also discusses the evolution of mosaic copy number alterations in healthy tissues.
The mother’s microbiome helps in the formation of the placenta (Pronovost et al., Science Advances). The authors of this paper treated pregnant mice with antibiotics that killed part of their gut microbiome and found that the placentas of antibiotic-treated mothers were smaller than those of mother mice with normal microbiomes. In addition, they had less-developed blood vessels needed to transport nutrients, oxygen and immune molecules from the mother to the fetus. This means that the gut microbiome plays an essential role in the formation of the placenta (in mice at least), further strengthening the link between mother’s microbiome and fetal development.
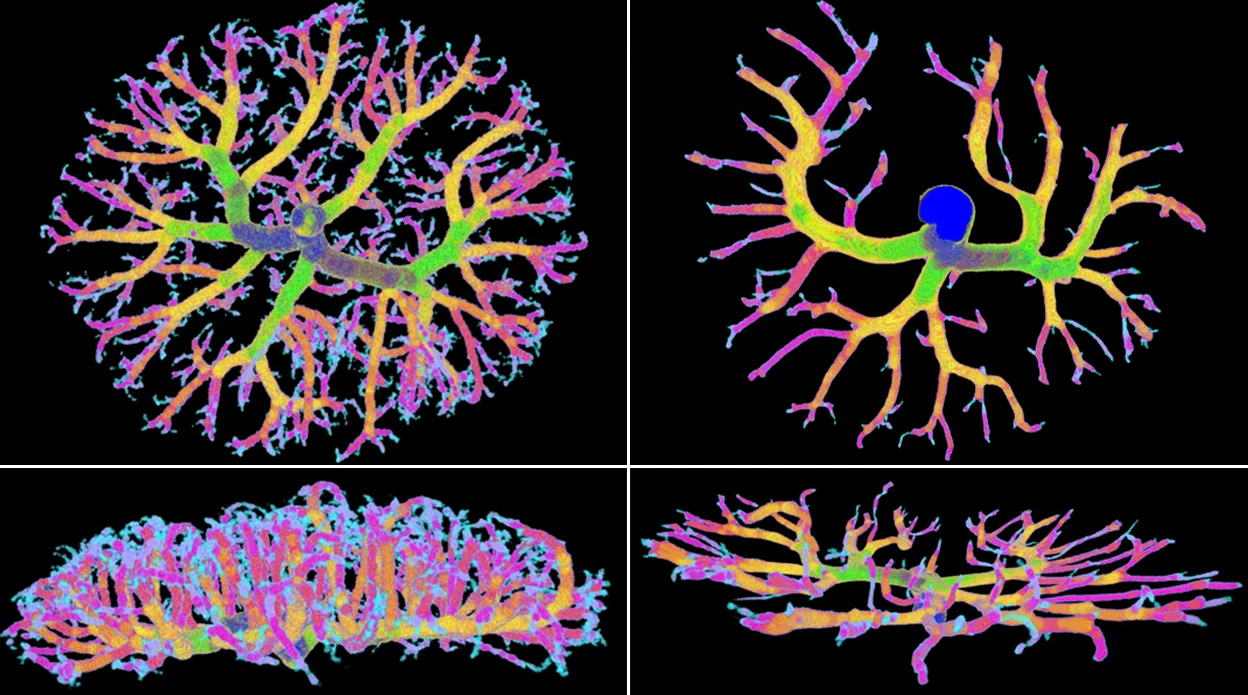
7. The maternal microbiome promotes placental development in mice, a short and beautiful experimental Science study, accompanied by a short and beautiful Nature explainer. Identification of cell states altered in disease (Dann et al., Nature Genetics): This paper investigates the question of how to optimally utilize existing single cell atlases (especially data from healthy individuals) to identify cell states altered in disease, and whether matched case-control data is needed for this task. It compares the same workflow for disease-state identification using as healthy reference either i) an atlas dataset (easier to get, but less specific); ii) a control dataset (expensive and smaller, but very specific); iii) a combination of both of the above, where the atlas is used for latent space learning, and the control as reference for differential analysis. They find that the combined approach of using a reference atlas for latent space learning followed by differential analysis against matched controls consistently increases precision and leads to improved identification of disease-associated cells, especially with multiple perturbed cell types.
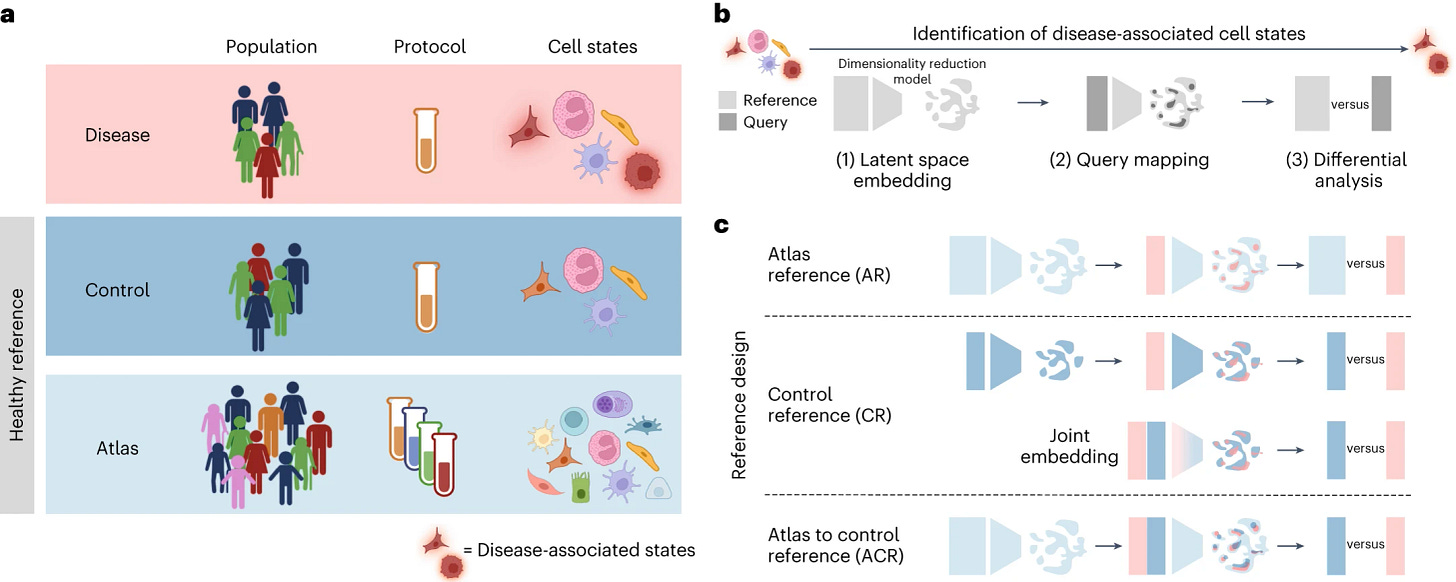
8. Precise identification of cell states altered in disease using healthy single-cell references, a new statistical methodology providing guidelines for designing disease cohort studies and optimizing cell atlas use. Resident regulatory T cells reflect the immune history of individual lymph nodes (Kaminski et al., Science Immunology). Regulatory T cells (Treg) are present in lymphoid and non-lymphoid tissues where they restrict immune activation, prevent autoimmunity and regulate inflammation: while Treg in non-lymphoid tissues are resident, those in lymph nodes (LNs) are considered to recirculate. In a beautiful series of experiments, this paper uses in situ labeling with a stabilized photoconvertible protein to uncover turnover rates of Treg in LNs in vivo. It finds that, similar to conventional T cells, Treg can form resident memory-like populations in LNs after adaptive immune responses, which might provide new therapeutic opportunities for the treatment of local chronic inflammatory conditions.
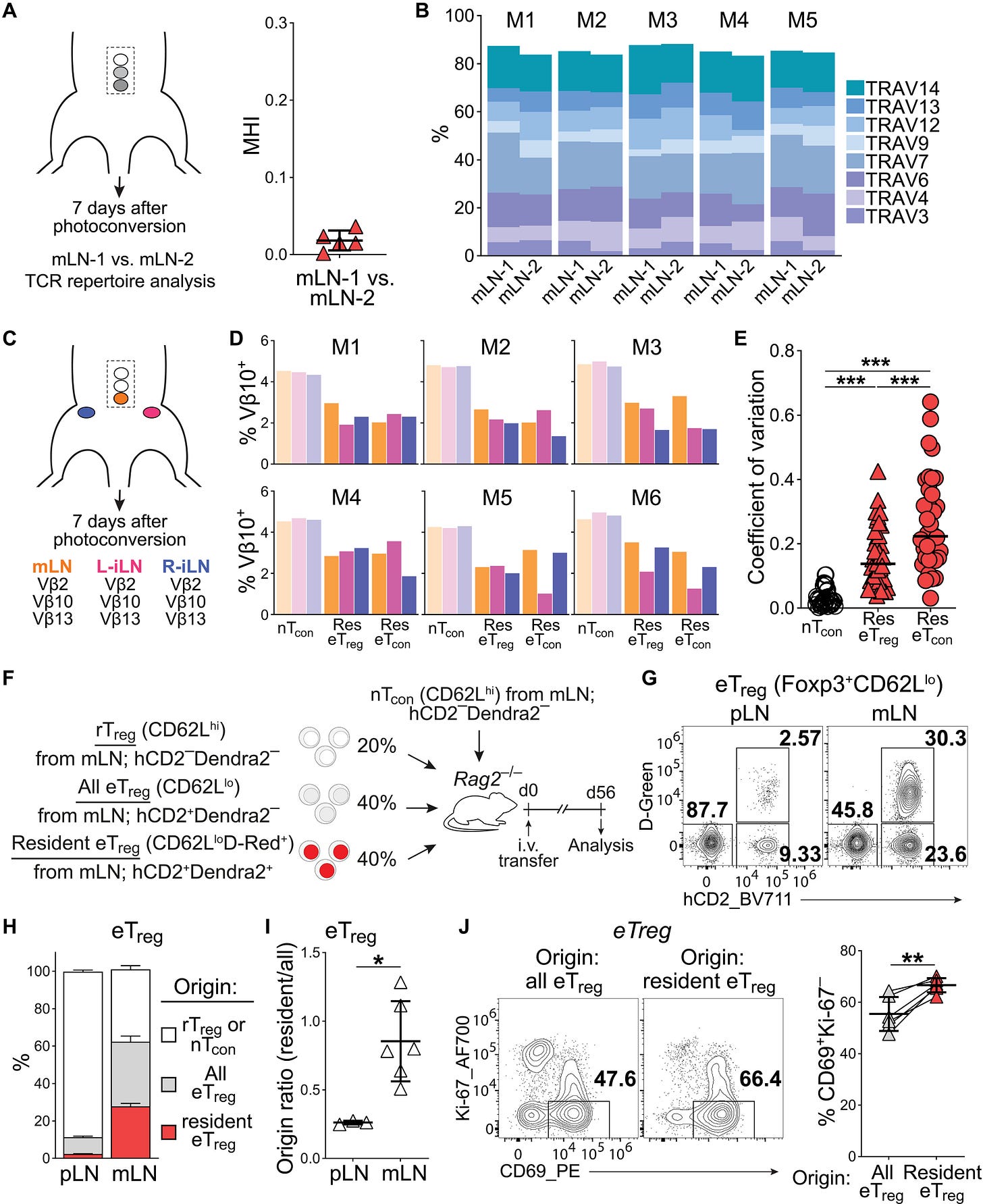
9. Resident regulatory T cells reflect the immune history of individual lymph nodes. This series of plots (Figure 7 in the paper) shows how each LN contains resident eTreg with exclusive TCR repertoires. The cancer-immunity cycle collection (Immunity). I don’t usually do Reviews in this newsletter, but this 7-article collection deserves an exception. The cancer-immunity cycle is a framework to understand the series of events that generate anti-cancer immune responses, introduced in 2013 by Chen & Mellman. Now, 10 years later, these review articles discuss progress and advances in the field, while highlighting key challenges: a telling example of our evolving understanding is the complex role of the tumor microenvironment in facilitating, not just suppressing, the anti-cancer response.

10. The cancer-immunity cycle: a collection of 7 review articles published in the journal Immunity marks 10 years since the concept was first introduced. These 7 reviews discuss in detail how our understanding of the role of the immune system in cancer initiation and progression has evolved, and the challenges than remain.
You made it till the end, that’s impressive. Here’s the bonus I promised.
That was 3x10 for October 2023. It you have suggestions of cool papers to include in November’s 3x10, please reach out either via Substack (Chat or Notes) or Twitter.



amazing work